TCR and BCR
sequencing services
Adaptive provides a sensitive and quantitative solution for T-cell receptor (TCR) and B-cell receptor (BCR) sequencing that helps you discover the breadth and depth of the adaptive immune system. Adaptive’s bulk TCR and BCR sequencing technology focuses on the CDR3 region—the most diverse part of an immune receptor—which determines antigen specificity and enables the adaptive immune system to recognize and respond to new threats. From experimental design to publication-ready data, this solution gives you the power to decipher the complexity of the adaptive immune system, providing fundamental insights into the body’s response to disease and therapy at the cellular level.
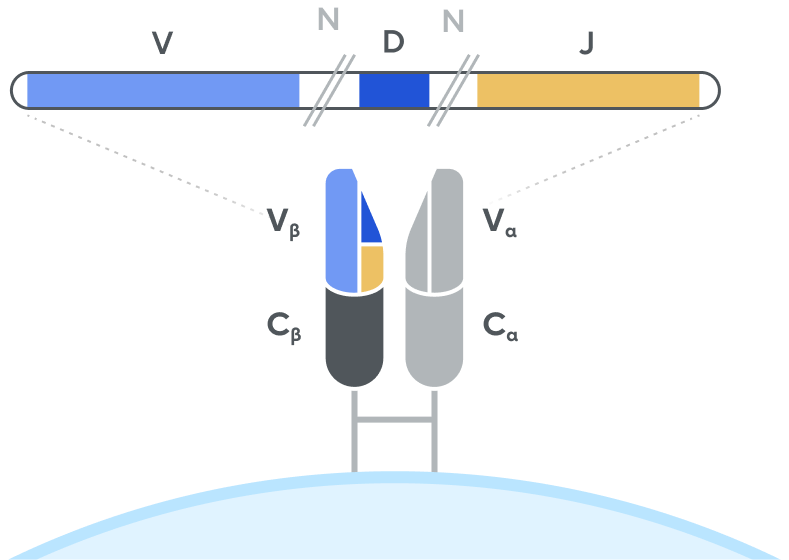
T-cell receptor (TCR)
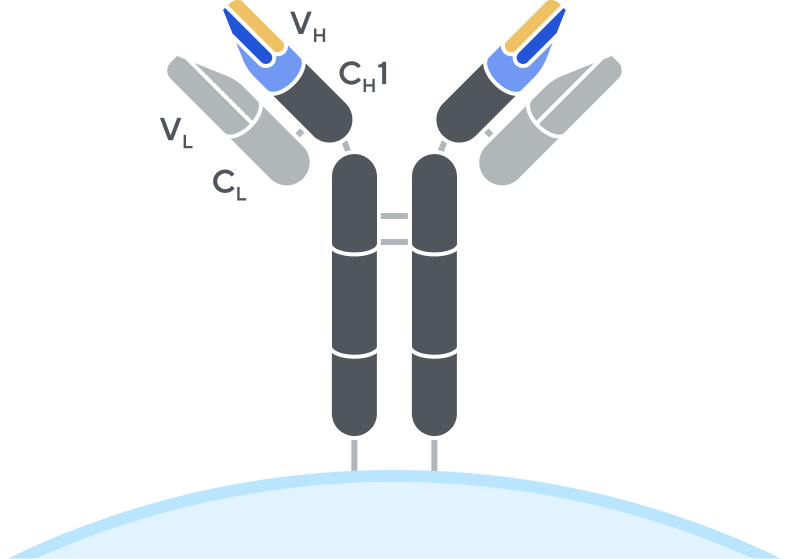
B-cell receptor (BCR)
Accuracy and quantification by design
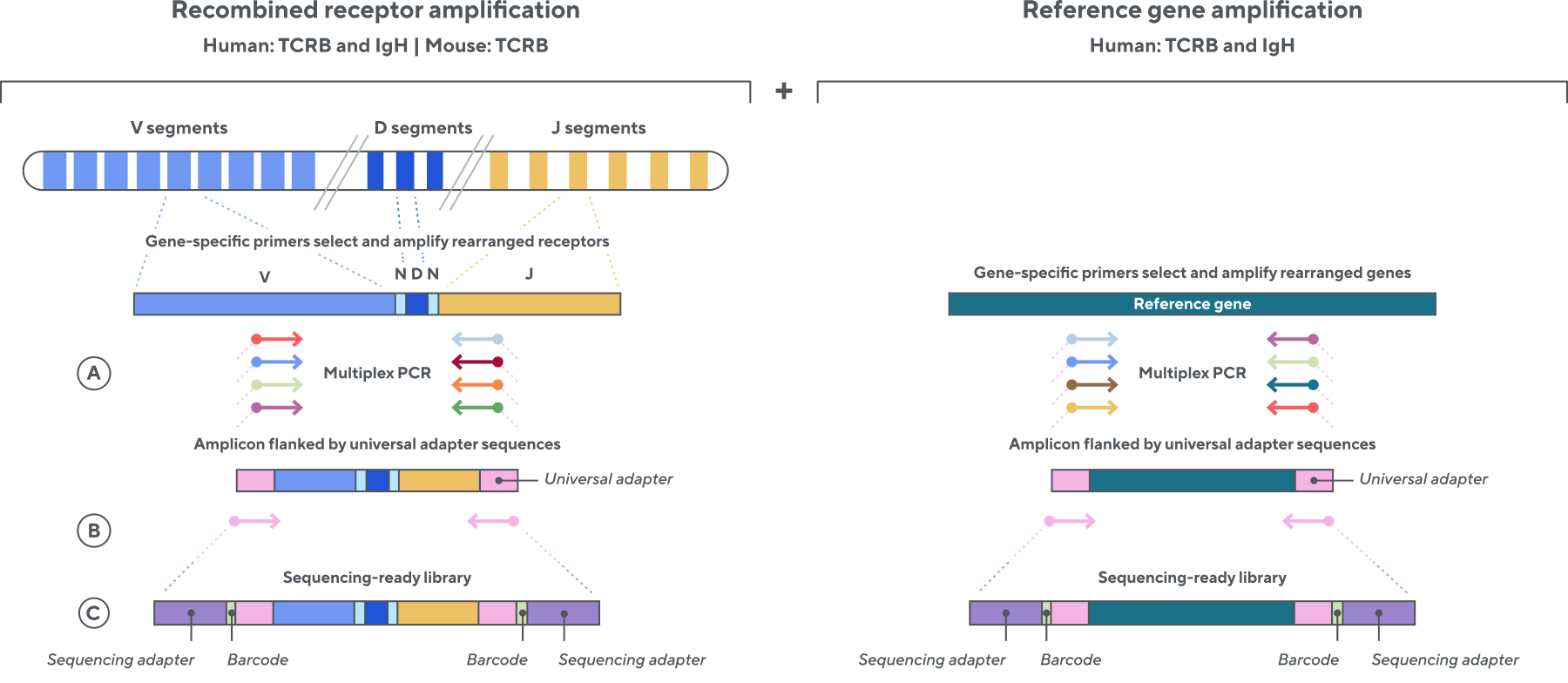
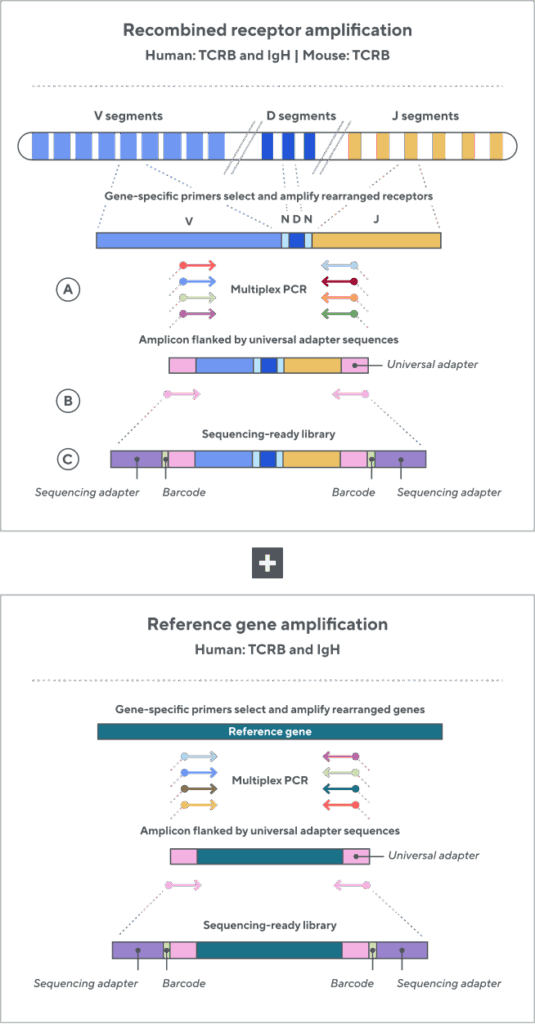
Adaptive’s TCR and BCR sequencing analyses use a proprietary multiplex polymerase chain reaction (PCR) assay to sequence directly from recombined genomic DNA (gDNA). We provide accurate, precise, and quantitative abundance data on your T- and B-cell populations—unlike the relative abundance or variable expression data of RNA-based assays. The assay contains rigorously designed synthetic immune templates as in-line controls, plus optimized primers that ensure accurate, quantitative, and unbiased results with batch-to-batch consistency. These proprietary features enable Adaptive’s bulk TCR and BCR sequencing assays to accurately quantify the immune repertoire over time, across people, and across studies.
True diversity is about seeing more of the repertoire
Adaptive’s high-throughput TCR and BCR sequencing analyses are designed to deeply sequence the immune receptor repertoire via the T-cell receptor beta (TCRB) and immunoglobulin heavy-chain (IgH) loci, respectively, to deliver the depth and breadth needed to truly capture immune repertoire diversity. High-throughput sequencing of the CDR3 variable chain of T-cell receptors and B-cell receptors provides powerful insights. The CDR3 sequence acts as a unique tag for each clonal lineage, which enables characterizing the diversity of the immune repertoire, tracking T cells and B cells over time, and measuring T-cell and B-cell responses due to disease and treatment. Detection and quantitation of rare or low-abundance clones is important in oncology, autoimmune, and infectious disease settings, and with Adaptive’s high-sensitivity TCR sequencing and BCR sequencing analyses, clonal expansion or contraction of these low-abundance clones is easily detectable and quantifiable.
Genomic DNA vs RNA or cDNA
Adaptive’s T-cell receptor and B-cell receptor sequencing assays are DNA-based and sequence from gDNA or RNA-derived complementary DNA (cDNA). The gDNA-based nature of Adaptive’s assays offers several advantages. From a workflow perspective, gDNA may be preferred, especially in archival specimens, as gDNA remains more stable compared to messenger RNA (mRNA). From an analytical and quantitative perspective, Adaptive’s proprietary sequencing approach quantifies each TCRB and IgH template, which, when using gDNA, provides accurate and precise T- or B-cell counts. This approach makes it possible to accurately assess clonal expansion and tissue density of T and B cells. In contrast, measurements from cDNA or mRNA are confounded by expression levels and may not provide an accurate measure of clonality or clonal expansion.
When gDNA is interrogated with Adaptive’s unbiased and quantitative assay, the output includes:

Template quantitation for ech TCBR or lgH sequence

T-cell or B-cell fraction

Absolute cell counts

Accurate repertoire metrics