TCR sequencing applications
Adaptive’s T-cell receptor (TCR) deep sequencing analyses provide a sensitive and quantitative solution that helps researchers, drug developers, and pharma partners discover the breadth and depth of the T-cell repertoire to advance the next generation of therapies. The assay is available for mouse and human samples, enabling researchers to begin development in preclinical studies and to continue observation as insights are translated into clinical settings.
Use in the drug discovery and development process
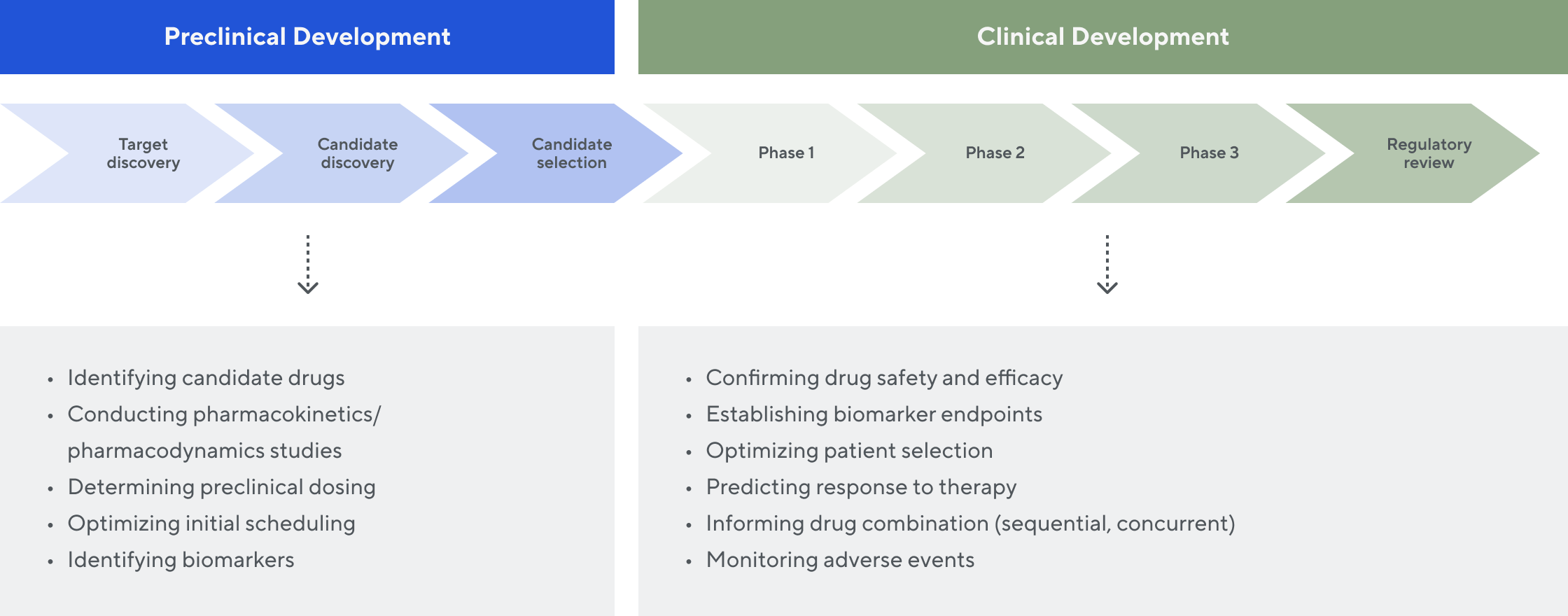
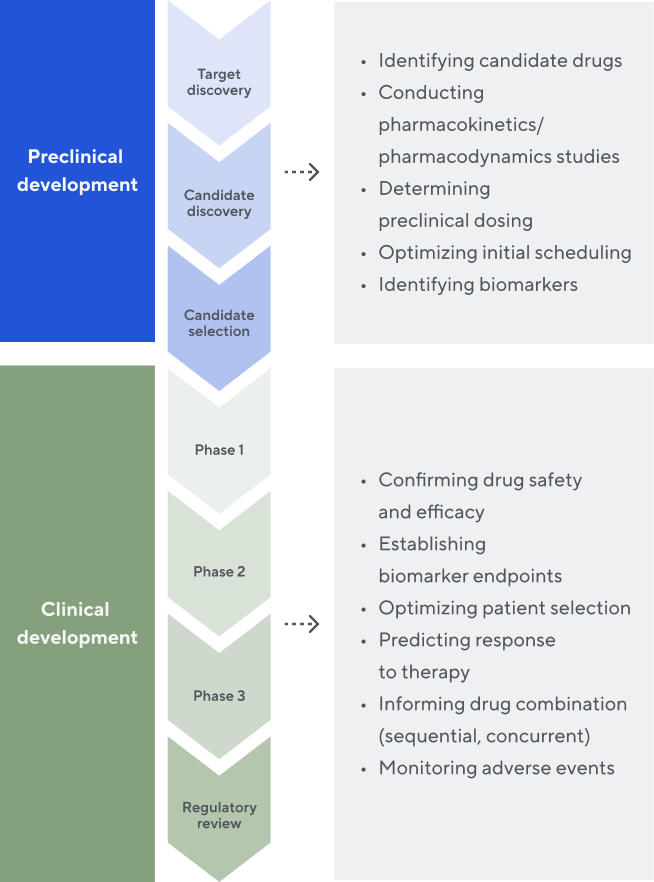
We work with our partners across the research and development value chain. The rich immune receptor data we generate provides valuable insights to inform and accelerate preclinical and clinical drug development—potentially saving significant cost and time.
Key metrics of TCR sequencing data
Adaptive’s synthetic immune repertoire and sophisticated bioinformatics provide unparalleled accuracy and precision in quantitatively assessing the T-cell repertoire. This exquisite accuracy enables precise quantitative of individual clone frequencies, clonal expansion, and assessment of key repertoire metrics.
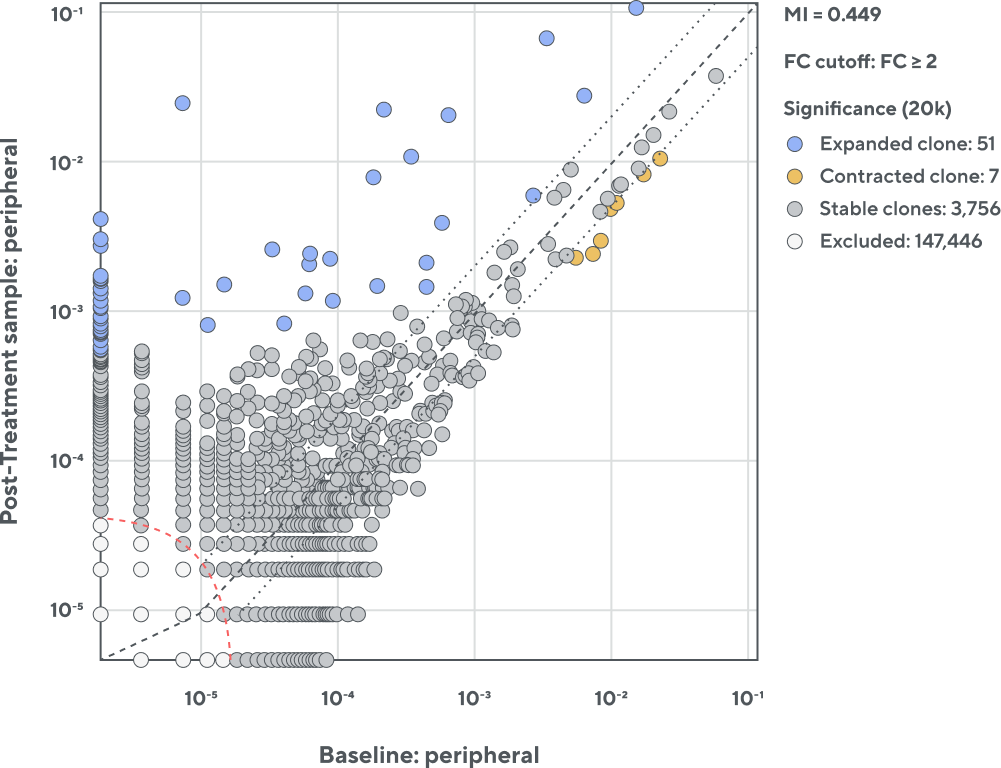

Expanded clone
Statistically significant greater frequency in post-treatment sample
Contracted clone
Statistically significant greater frequency in pre-treatment sample
Stable clones
Clones with no significant changes in frequency; increased scatter at low frequencies is due to sampling from a diverse pool of TCRs
Clone tracking
Track T- or B-cell receptors from diverse samples, pre- and post-treatment and over time, identifying and quantifying clonal expansion and contraction.

Repertoire properties
Quantify clonality and diversity of the T- and B-cell receptor repertoire.

Public clones
Identify shared T- and B-cell receptors.

Immune repertoire overlap
Detect clonal overlap between different samples or across different time points.

Lymphocyte count
View and monitor changes in T-cell fraction and B-cell fraction over time or between samples.

Mapping
Map T-cell signatures of exposure to repertoires.

Clone tracking
Track T- or B-cell receptors from diverse samples, pre- and post-treatment and over time, identifying and quantifying clonal expansion and contraction.

Repertoire properties
Quantify clonality and diversity of the T- and B-cell receptor repertoire.

Public clones
Identify shared T- and B-cell receptors.

Immune repertoire overlap
Detect clonal overlap between different samples or across different time points.

Lymphocyte count
View and monitor changes in T-cell fraction and B-cell fraction over time or between samples.

Mapping
Map T-cell signatures of exposure to repertoires.
T-cell signatures of exposure
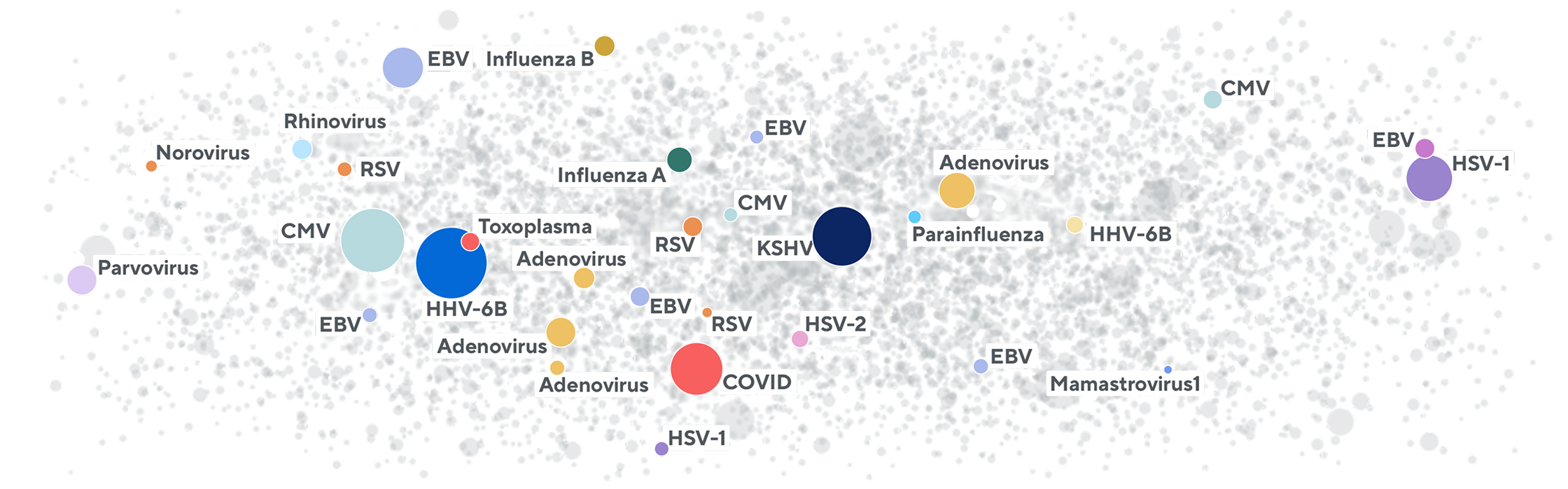
Adaptive hosts the world’s largest database of T-cell receptor sequences and is applying it toward matching TCRs to antigens, common exposures, and human leukocyte antigen (HLA) associations. This database contains T-cell receptor beta (TCRB) sequences with high-quality annotations for their specificity to HLA, exposures, and antigens. Adaptive has generated and validated these annotations over 10+ years of research using proprietary experimental methods and machine learning. The database is continuously expanding and available to partners to help them better understand their TCR sequencing data in the context of disease and treatment.
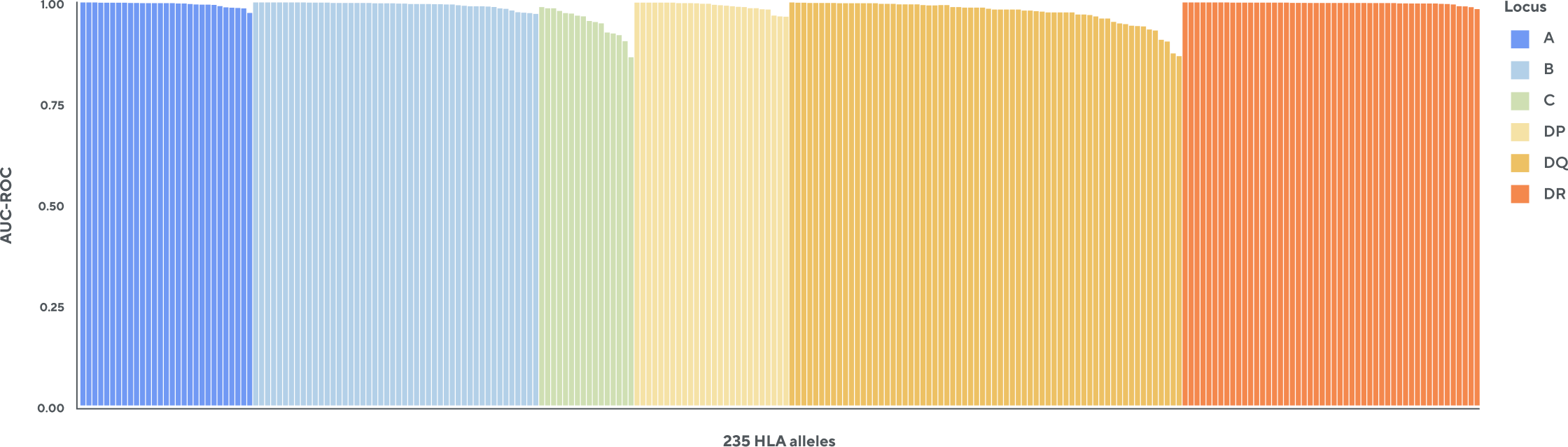
HLA typer infers the presence or absence of over 200 Class I and Class II HLA alleles
Foreign antigens are presented to T cells by protein complexes called “major histocompatibility complexes” (MHCs) that are coded by HLA genes. HLA type plays an important role in driving T-cell selection and in shaping T-cell repertoires. The importance of a person’s HLA type in transplant research is well understood, as are the associations of specific HLA alleles to common diseases. New research is emerging that explores the link between HLA and disease susceptibility, response to immunotherapy, and more. By adding HLA typing to samples processed using TCR sequencing, researchers can unlock new understanding of immune repertoire dynamics in the context of HLA type.
Adaptive’s T-cell receptor sequencing analyses can infer a sample’s HLA type based on the TCRB repertoire. The HLA classifier uses machine learning to provide a positive or negative call for over 200 Class I and Class II HLA alleles. This unique tool is based on TCR sequencing data generated from Adaptive’s partnership with Microsoft from approximately 4,000 HLA-typed samples.